RRSG Questionnaire 2019 - Summary
Many thanks to all who took the time to participate and to provide your feedback. It’s clear that the ISMRM community, while it may not be hanging from quite such a high precipice as more statistics-driven fields, also has its fair share of reproducibility problems.
This is probably the main take-home message from the whole survey:
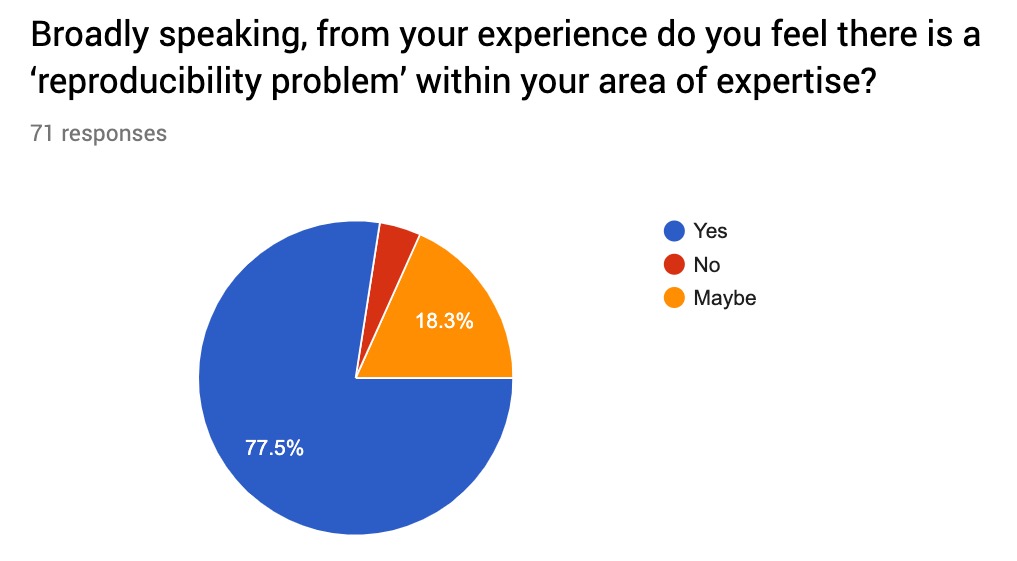
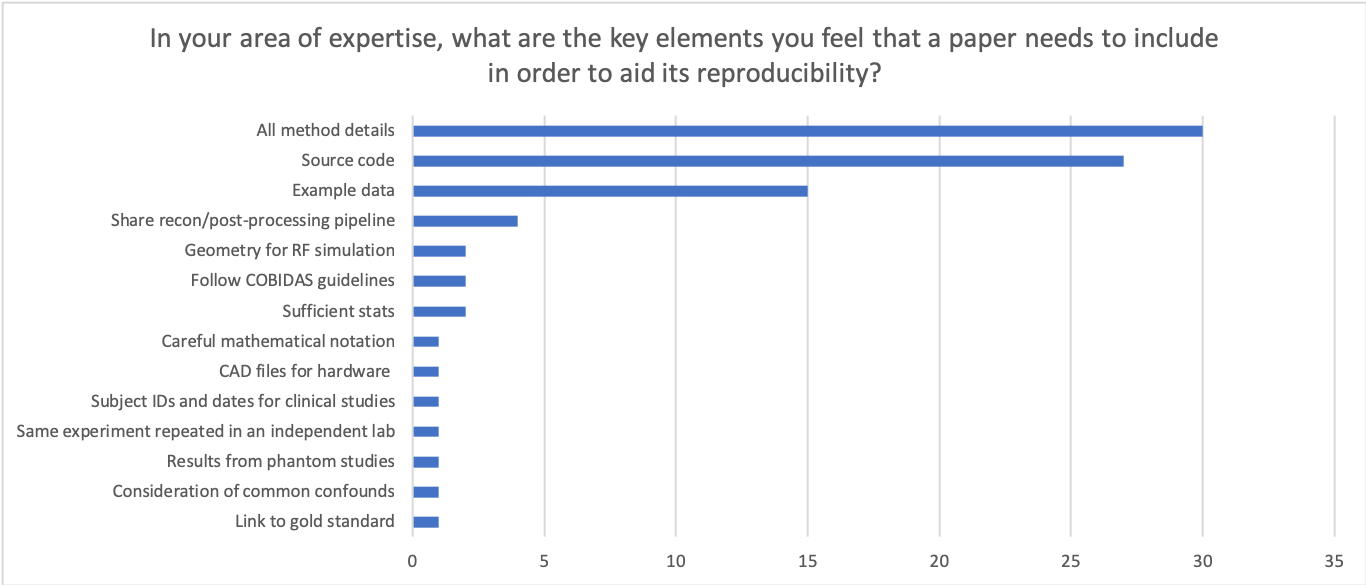
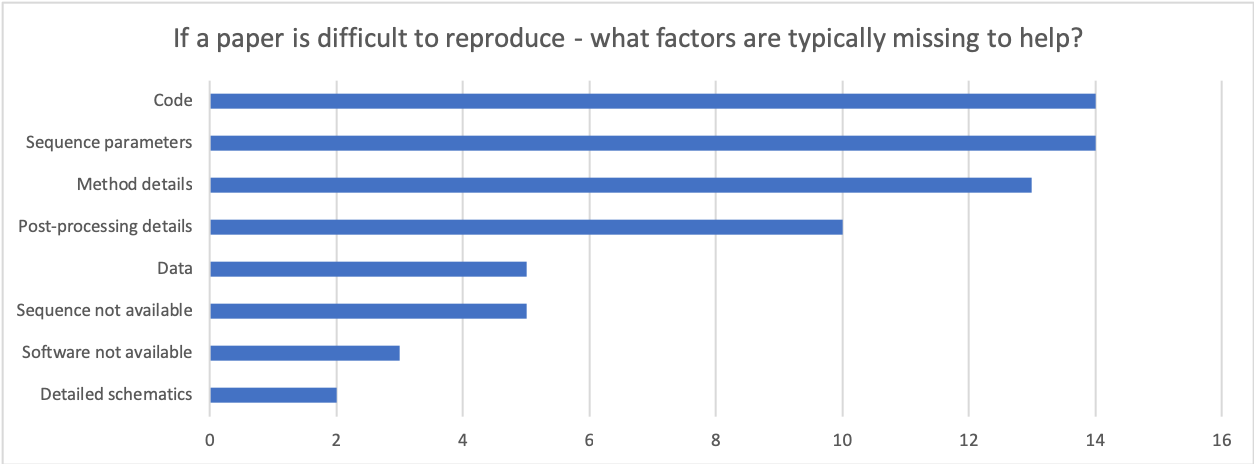
The results from the rest of the survey highlight that there are a number of factors that could be improved within our field - ranging right through from the hardware itself (e.g. attempting to match pulse sequences across vendors), image reconstruction algorithms, image processing pipelines and validation of new methods in representative cohorts.
My favourite quote from all the responses - which also sums things up quite nicely:
"Data and code or it didn't happen"
Browse full responses
You can read through all the answers provided to the survey by clicking here.
Summary of responses
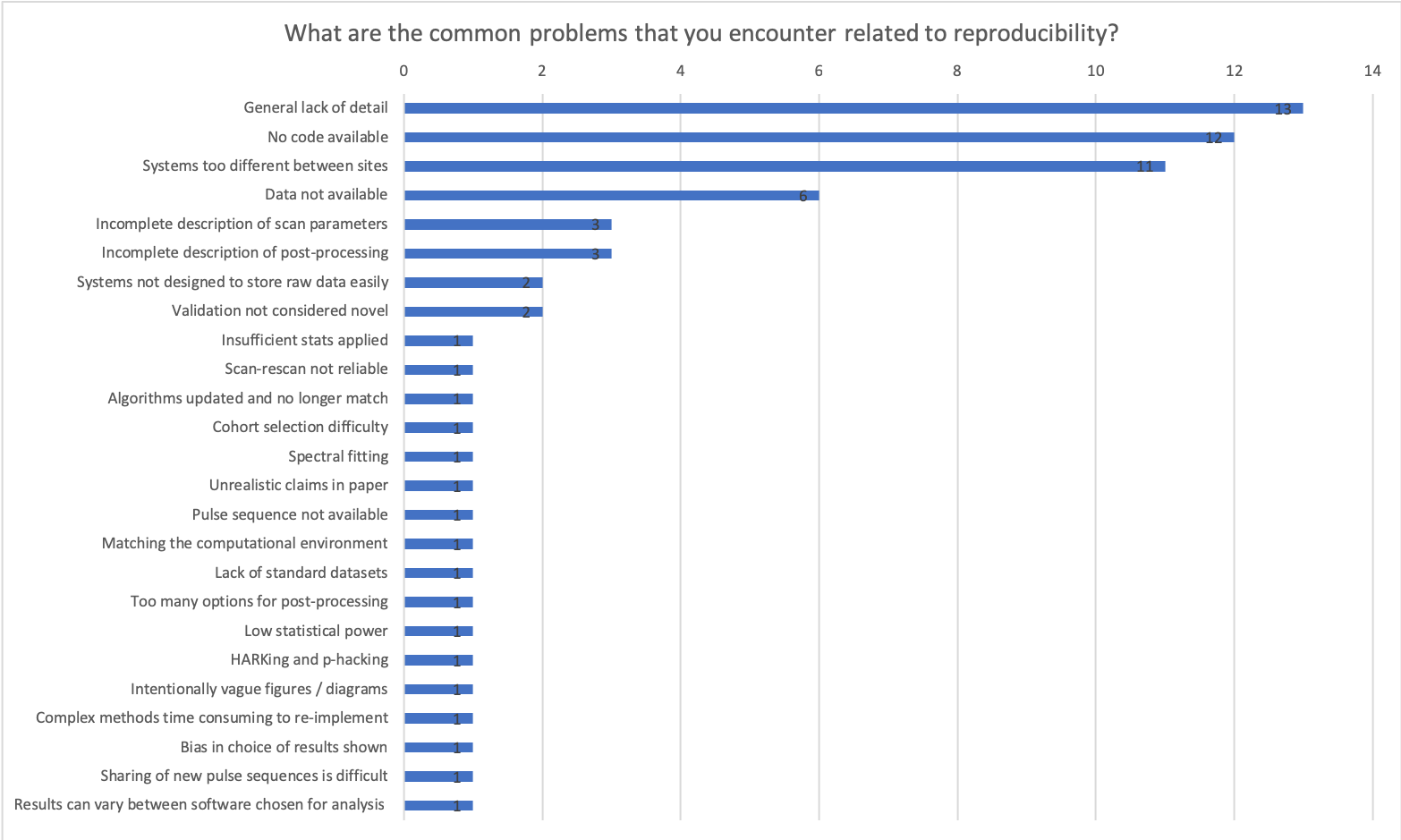
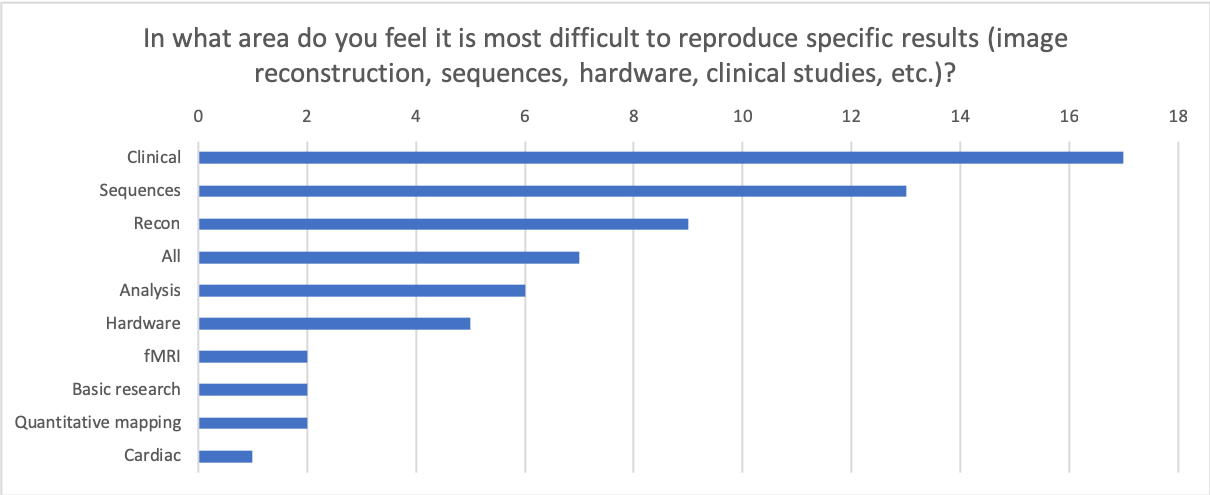
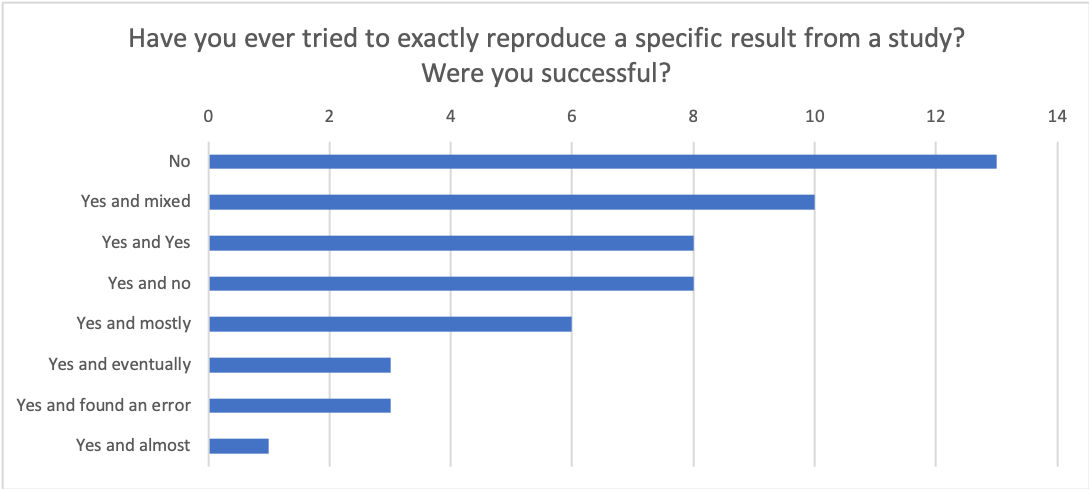
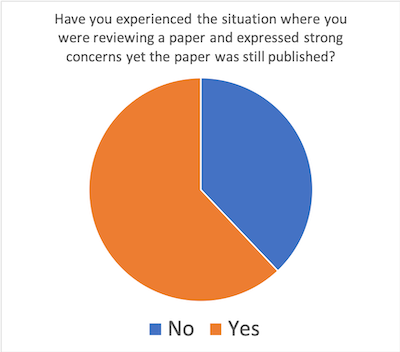
If so, what could have been changed to improve the process?
- Journals and funding agencies should make the importance of reproducibility more clear
- Manuscripts could be published with reviewer comments
- If authors say ‘code will be published’ there should be some check
- Need for a database that check if paper is already published elsewhere
- Access to code and data for reviewers
- Avoid publishing in Open-access journals!
- Editors need to be willing to make difficult decisions
- Editors should ensure reproducibility, e.g. if result can only be obtained with proprietary hardware
- Data should be supplied to allow testing of statistics reported
In the area of your expertise, are there data sharing initiatives you are aware of? (please provide links if so)
- https://openneuro.org/
- Some IMI projects (imi.europa.eu) are quite active in datasharing
- Yes, major NIH funded neuroimaging probjects (ADNI, ABCD, HCP,…)
- ISMRM, Multiinstitutional
- http://www.opensourceimaging.org
- 1000 functional connectomes. Human connectome project
- http://mridata.org, segmentation challenges, github repos
- http://ninaweb.hevs.ch/
- NITRC
- MyoSegmentum: https://osf.io/svwa7/?view_only=c2c980c17b3a40fca35d088a3cdd83e2
- https://zenodo.org/
- Nci cancer imaging archive
- ADNI, human connectome
- datalad
- neurovault
- DiPy, various MATLAB tools…
- INDI, ABIDE, ADHD200, HCP, CAMCAN,
- several, e.g. OASIS, ADNI. most clinical consortia need to share their data ultimately
- https://researchdata.ands.org.au/
- I am aware of databases such as ADNI and PPMI,))
Are you aware of reproducibility initiatives in other fields that we might not be aware of that could help direct the RRSG efforts? (please provide links if so)
| CAMARADES Systematic Reviews | https://www.nc3rs.org.uk/camaradesnc3rs-systematic-review-facility-syrf |
| NYU fastMRI dataset | https://fastmri.med.nyu.edu/ |
| Most new ML/DL conferences highly encourage code sharing. NeuRIPS, CVPR, etc | |
| Journal of Open Source Software | http://joss.theoj.org/ |
| In genomics and bioinformatics, a lot of work has been done on Nextflow (pipeline engine enabling reproducibility if correctly used, www.nextflow.io) and on Singularity (container system: https://singularity.lbl.gov/) | http://www.nextflow.io |
| German National Cohort | https://nako.de/informationen-auf-englisch/ |
| UK Biobank | https://imaging.ukbiobank.ac.uk/ |
| THPC reproducibility iniative, (TECHNICAL CONSORTIUM ON HIGH PERFORMANCE COMPUTING), | https://tc.computer.org/tchpc/home-page/reproducibility/ |
| We are working on image processing reproducibility in ExploreASL | https://sites.google.com/view/exploreasl |
| Provision of help for MR users creating a local normal range properly (because reproducibility is a problem). See https://doi.org/10.1002/jmri.26683 and https://bit.ly/2XE1Ngk | https://doi.org/10.1002/jmri.26683 |
| International Brain Initiative | https://www.internationalbraininitiative.org/news/incf-brings-international-brain-initiatives-together-align-systems-and-data-standards |
| NIH Rigor and Reproducibility | https://www.nih.gov/research-training/rigor-reproducibility |
| SfN - Enhancing Rigor | https://neuronline.sfn.org/Collections/Promoting-Awareness-and-Knowledge-to-Enhance-Scientific-Rigor-in-Neuroscience |
| Collection of papers on Human Brain MRI reproducibility | https://www.nature.com/collections/yglmshrkbg |
| Neuron article: "The Costs of Reproducibility" | https://www.sciencedirect.com/science/article/pii/S0896627318310390 |
What software tools are you using in your research? Are they open source?
Note of course that the ‘user count’ above refers only to counts in the questionnaire responses, and isn’t a real guide to the popularity of the software…!
What specific software tools / resources do you feel are missing?
- guidelines on minimum standards of study design and reporting of in vivo (animal/human) MR that journals and peer-reviewers could refer to.
- Proper data sharing repositories with specified/controlled access
- A registration algorithm for noisy/undersampled data, that does not just work for brain images. (ANTs toolbox is hard to install with a general lack of documentation)
- Sequence building tool
- Analysis tools
- FPGA programming
- Tools for data IO/conversion from different vendors to open formats!!!
- Advanced image reconstruction, QSM, although some are available . Writing help for such tools is hard, but very helpful
- Shareable, vendor-idependent sequence programming dicom processing tools
- code and data repository, approved/accepted by industry
- Sequence development tools that integrate seamlessly with existing scanners, provide feature parity with the vendor supplied ones, and that allow us to share and openly discuss sequence implementations without signing non-disclosure agreements.
- Good tools for visualizating and checking the quality of big datasets. I believe it is too easy to just start processing and creating results, while some of the results might just be caused by artefacts or just spurious correlations. Visualization would help with this.
- Sometimes it is difficult to know/reproduce exactly what the scanner is doing in image reconstruction but of course proprietary info
- quantitative analysis tools; accurate lesion segmentation
- software support/development is lacking in some of the packages above
- tutorials and learning for beginners to get them up to speed on code sharing
- usually reproducibility is not tested, we are doing this now
- Cloud-based or browser-based medical image processing is not commonly available
- A culture of citing code via DOIs as well as citing academic references would provide a better incentive and a way for people to be acknowledged for their work theme specific standardized tools (ultra-high res fmri ), but this is work in progress within the ultra-high res community
- I think that when a novel sequence is described in a paper, it happens that details are not sufficient to reimplement the software needed to process the data and reproduce the results. It is a choice of the authors, often.
- Decent collaborative scientific editing software
Do you have any suggestions for initiatives you would like the RRSG to pursue?
- initiatives towards vendor-neutrality. Harmonization efforts which can allows researchers to replicate previous work on a different vendor’s unit and allow multi-site clinical studies or data sharing between groups that don’t share similar vendor machines. Especially given the push to AI, most of the network trained on data from one vendor’s system may not quite applicable on data from another vendor - expect perhaps with transfer learning
- guidelines on minimum standards of study design and reporting of in vivo (animal/human) MR to facilitate reproducible research
- define a standard format in which code and data associated with MRM and JMRI papers can be shared and connecting to the publishing committee in order to include this in the review / publishing routine. An browser based platform to reproduce figures from papers would be amazing.
- Might be outside scope of what the study group can accomplish, but it would be nice if the MRI journals required/encouraged authors to share the details of their work in supplementary information and code repositories.
- development of standard libraries to compare results to. For example we can identify a CS reconstruction or a registration software as our gold standard and every new study includes a comparison with respect to these techniques.
- Publications facilitating the reproducibility should be incentivized. Also, Hardware really needs a push.
- Make the ISMRM journals, MRM and JMRI, not accept any paper submissions without code or data access, or open an extra journal for reproducible MR research!!!
- Education courses for methodologists and scientists running studies on how to make sure their work and results are reproducible.
- SOP , guidelines, education
- ISMRM should promote effort to store, preserve, and share raw data acquired from MRI for research purposes
- Industry support for reproducible research
- Getting faculty and senior people on the same page about RRSG would be very helpful. Even if I wanted to make my code more readable or spend time writing up details on how to implement the method, if my PI doesn’t think it’s a useful way to spend my time it won’t get done. The only times I’ve uploaded code to github are when reviewers specifically asked for it
- I think we should bring some people from the industry to our side. We have to get the big vendors on board, and get them to open source at least part of their software or write standardized APIs so that we can safely manipulate the scanner without using their software.
- Good practice manual? Workshop?
- I would like to use python more for image reconstruction but most tools are in Matlab and it is hard to shift I think there is a ISMRM raw data format but the vendor raw data format can be a barrier to sharing. Perhaps there is a tool to convert from vendor to ISMRM format. Often the vendor file has esoteric fields so I suppose a decision has to be made on which parameters to keep or rename.
- In addition to items noted above, I see several issues that affect the reproducibility of biomedical MR experiments: insufficient knowledge of physiology and insufficient knowledge of statistics and experimental design. I would love to see these topics on the program
- encourage full reporting of MRI parameters & back reporting if possible on published work via supplements; an open source database of sequence parameters used which is linked to papers and searchable
- I think the RRSG could try to collaborate with other study groups (e.g., work with one at each ISMRM scientific meeting) to increase awareness and help guide initiatives to solve issues specific to the partner SG (e.g., hardware reproducibility –> Engineering SG).
- specific papers on reproducibility of a certain method
- Currently only the part from submission to publication is peer-reviewed. It would be great to set up a way to also peer review the steps leading to the paper: e.g. the study protocol, the acquisition chosen, image processing etc. This will not only increase reproducibility, but also the medical-ethical value of the patients that are scanned: their research contribution will have more value.
- Reproducibility can be increased by standardisation of protocols at each stage - data acquisition, processing and analysis. I would like to see more standardised open source software for image processing and analysis for all areas of MR research (not limited to a particular organ or MR technique)
- as I wrote above but not sure how to achieve it! I think MRM in general should enforce its stated guidelines for use of git (or similar) with SHA quoted for much more work.
- Better sharing of sequences.
- I think an initiative to reproduce results from a paper of choice (without constraints on the journal) would be very interesting
- Template text for patient ethics to permit data sharing. Template data sharing agreements (on the assumption it can’t be truly open). Example settings for data anonymisation software.
- Most important is I think to change the culture, especially at the top, to not only promote open science, but make it an essential activity, valued and rewarded with clear career benefits (notably through changes in funding system) fundraising for implementing consensus methods with all 3 main MR vendors.
