In April 2019 the RRSG challenged the ISMRM community to reproduce the results from a seminal paper in the field. The goal of this initiative is to select seminal papers from our field, and ask the community to reproduce the core findings/algorithms/implementations from these papers. The main motivation behind this initiative is to:
- Over time, create a library of standard reference implementations that can be used for comparison when publishing new methods.
- The ISMRM is currently creating educational content by curating educational lectures from past annual meetings and assembling them into online courses (https://www.ismrm.org/online-education-program/). An accompanying repository of reference implementations would be valuable additional content that the ISMRM research community could provide.
- Compare the individual submissions in terms of consistency of results, computation time and hardware/programming language requirements. Note: This is not to be intended to be a “ranking” of the submissions as would be done in a competition, but as an assessment of their variability.
- This initiative can be a great opportunity for students and trainees to gain additional visibility, especially if they come from smaller labs and countries where they do not have the opportunities to go to every ISMRM meeting and workshop to present their work to the research community.
The detailed submission instructions can be found here for reference.
The paper
The paper selected for this challenge, which participants were asked to reproduce:
Klaas P. Pruessmann, Markus Weiger, Peter Börnert, Peter Boesiger. Advances in sensitivity encoding with arbitrary k-space trajectories. Magn Reson Med. (2001); 46(4):638-51.
The data
We provided two example datasets, brain (12 receive channels, 96 radial projections) and cardiac (34 receive channels, 55 radial projections), from a radial trajectory acquired with multi-channel coils. The data is provided in the h5 format, and we are following the conventions of the BART toolbox regarding array dimensions of the raw data [1, Readout, Spokes, Channels] and the trajectory [3, Readout, Spokes] where the first dimension encodes the k-space coordinate (for 2D acquisitions the third coordinate is always zero) and the unit of measurement is 1 / FOV.
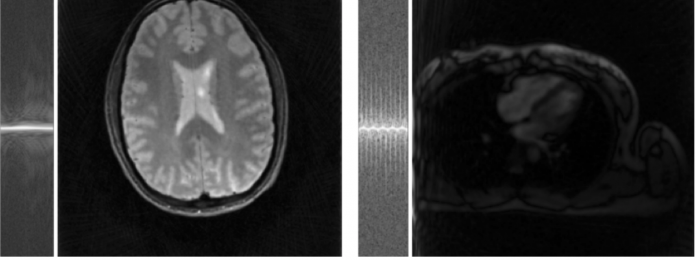
Brain data (5.3 MB): rawdata_brain_radial_6proj_12ch.h5: rawdata: [1, 512, 96, 12], trajectory: [1, 512, 96]
Cardiac data (5 MB): rawdata_heart_radial_55proj_34ch.h5: rawdata: [1, 320, 55, 34], trajectory: [1, 320, 55]
We also provided starter-scripts for MATLAB and Python which are able to read in and display the data. These scripts are also available from the Github repository.
Submissions
Collated list of submissions - May 2019:
| Authors (or principal author) | Link | Info |
|---|---|---|
| Steven
Baete (NYU) |
https://bitbucket.org/sbaete/ismrm2019reprodcgsense | MATLAB, NUFFT (Fessler), gpuNUFFT (Schwarzl, Knoll) |
| Alexander Fyrdahl (Karolinska Institutet and Karolinska University Hospital, Sweden) | https://github.com/fyrdahl/rrsg_challenge | MATLAB, NUFFT (Fessler) |
| Kerstin Hammernik (Graz University of Technology) | https://github.com/khammernik/ISMRM2019_RRSG | Python, BART, primal-dual-toolbox, medutils |
| Seb
Harrevelt (Eindhoven University of Technology) |
https://github.com/zwep/ismrm19_challenge | Python, PyNUFFT (Lin), |
| Namgyun Lee (University of Southern California) | https://drive.google.com/file/d/10qD6K-sCEkNjpynRZTpLm8VBUPJFhJCt/view | MATLAB, custom MEX for gridding |
| Gilad Liberman (MGH) | https://github.com/giladddd/LinopScript | MATLAB, Demo of linear-operator scripting for BART on
challenge datasets |
| Michael Loecher (Stanford) |
https://github.com/mloecher/rrsg_challenge | Python, custom Cython for gridding |
| Oliver Maier (Graz University of Technology) | https://github.com/MaierOli2010/ISMRM_RRSG | Python, BART, requires GPU for use of GPyFFT |
| Franz Patzig, Lars Kasper, Thomas Ulrich, Maria Engel, Johanna Vannesjo, Markus Weiger, David Brunner, Bertram Wilm, Klaas Prüssmann (ETHZ) | https://github.com/mrtm-zurich/rrsg-arbitrary-sense | MATLAB, custom gridding in MATLAB |
| Ludger Starke (MDC-Berlin) |
.../reproducibleResearch19_LudgerStarke.zip | MATLAB, BART |
| Ye Tian (UCAIR - University of Utah) |
https://github.com/YeTianMRI/ISMRM-2019-reproducible | MATLAB, NUFFT (Fessler) |
| Ke Wang, Miki Lustig, Ekin Karasan, Suma Anand, Volert Roeloffs
(Berkeley) |
https://github.com/KeWang0622/rrsg_challenge_sigpy | Python, SigPy (Ong) |
